Data Analysis with Python
I decided to dedicate this page to some useful tutorials about analysis, manipulation, and visualisation of data in the tabular form using Pandas, Numpy, and Matplotlib libraries of Python programming language. Now that I am sending this post the world is going through a massive crisis caused by the COVID-19 pandemic and that is where the data analysis and machine learning predicting algorithms come to play and show its significance. Therefore, in order to go along with the current vibe, I am going to use some open source datasets for each presented example which covers different aspect of statistics regarding the pandemic. I use Jupyter Notebook for the code development as it provides an interactive environment and presents the tabular data in an organised and unique way. Before installing Jupyter notebook, the Anaconda distribution package of Python is better to be installed to remove the need of installing each library separately.
import pandas as pddata = pd.read_csv('covid_19.csv')data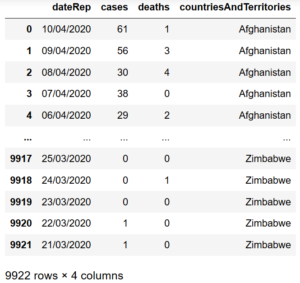
Now let’s see which countries are listed in this dataframe. To do that one just needs to choose ”countriesAndTerritories” columns and use the ”unique” method on it to get the name of last ten countries in the form of a list:
data['countriesAndTerritories'].unique()[-10:]then if we assign the above expression to a variable cou and get the length of it by passing it through the ”len()” function, we get to the number of listed country in the dataframe:
cou = data['countriesAndTerritories'].unique()
len(cou) then if we assign the above expression to a variable cou and get the length of it by passing it through the ”len()” function, we get to the number of listed country in the dataframe:
cou = data['countriesAndTerritories'].unique()
len(cou) Now to rank countries based on number of fatalities the dataframe needs to be groupedby based on column ”countriesAndTerritories” and use the ”sum” method to get the total numbers since the begining of pandemic.
data.groupby('countriesAndTerritories').sum()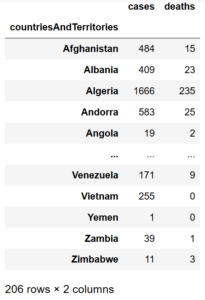
data.groupby('countriesAndTerritories').sum().sort_values(by='deaths',ascending=False)[:10]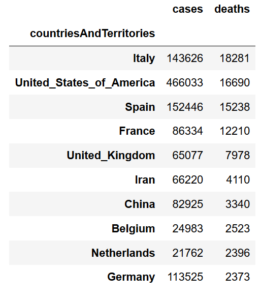
The ”countriesAndTerritories ” seems to be a long name for the column we are going to use its information many times so it seems reasonable to rename it to a shorter name such as ”countries” by applying ”rename” method on the dataframe as follows:
data.rename(columns={'countriesAndTerritories':'countries'},inplace=True)
data.columnsnote that the rename method took two attributions namely ”columns” and ”inplace”. A dictionary is passed for the first one contaning old and new names of the column we would like to rename, and the inplace took the boolean value of True to replace the new dataframe (contaning updated column name) with the previous version.
Taking a look at the name of countries, and following the fashion of using shorter names, one can change the ”United_States_of_America ” to the ”US” and ”United_Kingdom” to the ”UK” by applying the ”loc” method on the dataframe as follows:
data.loc[data['countries']=='United_States_of_America', 'countries']='US'
data.loc[data['countries']=='United_Kingdom', 'countries']='UK'basically by typing the above code, we tell the Python’s interpreter to locate the ”countries” columns of dataframe and if the value (rows) is equal to ‘United_States_of_America’ then change it to the ‘US’.
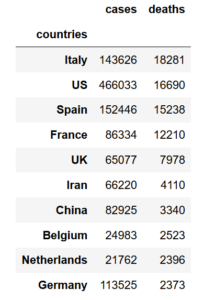
As the next step, it seems beneficial to add a column to the above dataframe outlining the death rate by dividing total numbers of deaths by the total numbers of cases. Thereby, if we apply the groupby method again similar to the way it is done before and assign a new variable (dataframe) we see the below image. Note that the second attribution of groupby method is to have the countries name as a seperate column and not as the index of the new dataframe.
df = data.groupby('countries',as_index=False).sum().sort_values(by='deaths',ascending=False)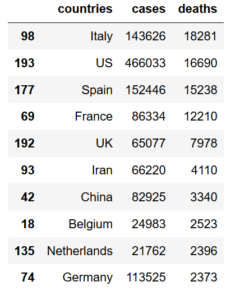
As you can see, the index of newly-defined dataframe ”df” belongs to that of old one ”data” and to reset it we just need to apply ”reset_index” method with an attributation to drop the old index as follows:
df=df.reset_index(drop=True)Note that without ”drop=True” attribution, the method keeps the old index as a seperate column which is not desirable in this case.
In order to add a new column named ”death-rate %” to the current dataframe the following needs to be implemented:
df['death-rate %'] = (df['deaths'] / df['cases']) * 100df=df.head(10)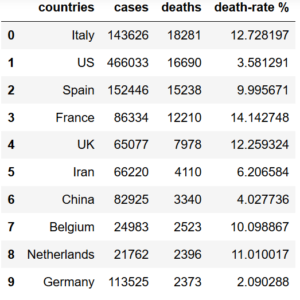
Using pyplot class of matplotlib library enables us to plot the above data in the form of bar charts where the horizontal axis specifies the country names and the vertical denotes numbers of cases, numbers of deaths, and death-rate percentage respetively.
from matplotlib import pyplot as plt
import numpy as np
fig, axe = plt.subplots(1,3,figsize=(20,5)) # adding three subplots in one row and three columns (1,3) in a figure with the size of 20*5
plt.subplots_adjust(wspace=0.2) # adjusting the horizontal space between subplots
colour = ['r','g','c'] # making a colour list to assign to the subplots
label = ['Infected Cases','Deaths','Death Rate Percentage'] # making label for the corresponding subplots
for i in range(3):
axe[i].bar(df.index,df.iloc[:,i+1], width=0.3, color=colour[i],edgecolor='k') # drawing a bar chart on each subplots. The index of dataframe (df.index) denotes the countries' name and df.iloc[:,i+1] selects the entire row for columns of 'infected cases','deaths', and 'death rate %' respectively.
axe[i].set_xticks(list(range(10))) #assinging 10 tickmarks to the horizontal axis on each subplot
axe[i].set_xticklabels(df['countries'], rotation=30)# assining name of countries as a lable to the tickmarks of horizontal axis
axe[i].set_ylabel(label[i]) # assigining labels to each subplot based on the label list defined before the loop
plt.savefig('barcharts.png',dpi=600,bbox_inches='tight') # saving the figure with a tight border in the png format and dpi of 600 pixel per inch
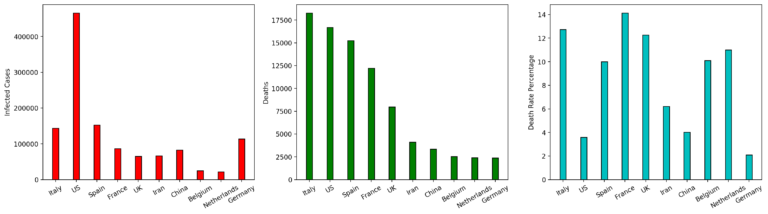
cou = list(df['countries'])cou_df=data.loc[data['countries'].apply(lambda x: x in cou)]cou_df_grouped = cou_df.groupby(['dateRep','countries']).sum().unstack()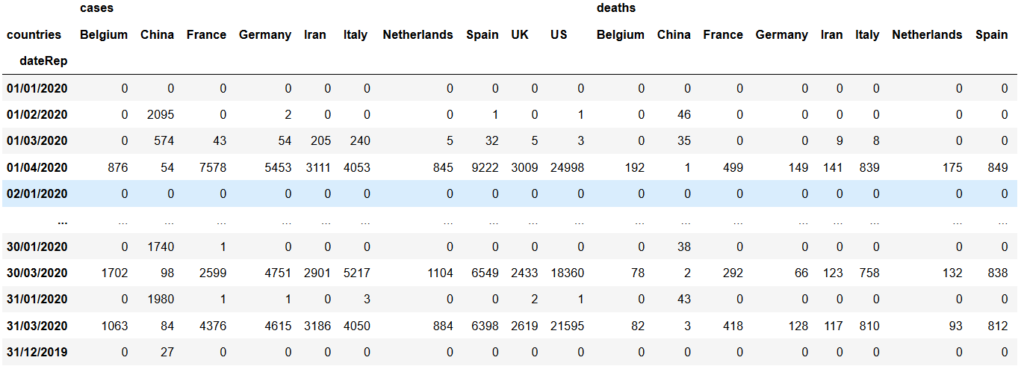
Note that the unstack() method makes the countries to appear as columns. We could also have the same arrangement by applying a pivot_table() method on the ‘cou_df’ dataframe but we stick with groupby() and unstack() methods for now.
Next step is to rearrenge the appearing order of countries (for both cases and deaths columns) from left to right in a descending manner based on the number of deaths according to the below list:

col=list(cou_df_grouped.columns)
first_index=[] #getting the index number of columns under the subcategory of 'cases'
second_index=[] #getting the index number of columns under the subcategory of 'deaths'
for i in range(10):
for elements in col:
if(elements[0]=='cases'):
if(elements[1]==cou[i] ):
first_index.append(col.index(elements))
elif(elements[1]==cou[i]):
second_index.append(col.index(elements))
cases_col=[]
deaths_col=[]
for i in range(10):
cases_col.append(col[first_index[i]])
deaths_col.append(col[second_index[i]])
rear_df = cou_df_grouped[cases_col + deaths_col]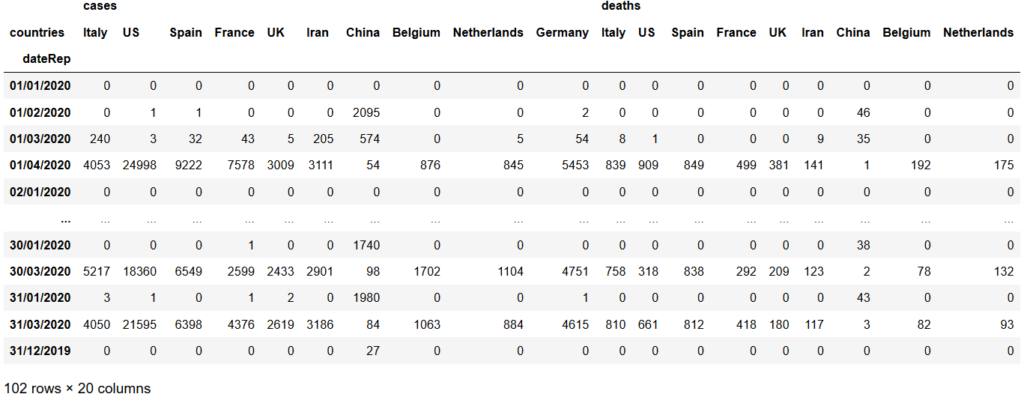
In order to be able to derive the total numbers of infected and dead cases the ‘cumsum’ method can be applied to calculate the cummulative summuation up untill each date:
rear_df_cum = rear_df.apply('cumsum')from matplotlib import pyplot as plt
import numpy as np
from scipy.interpolate import make_interp_spline
inter=[]
y_new=[]
inter_cum=[]
y_new_cum=[]
y1=0
colour = ['r','g','b','salmon','violet','gold','silver','lime','cyan','brown']
fig, axe = plt.subplots(5,2,figsize=(25,40))
plt.subplots_adjust(hspace=0.3)
x_new = np.linspace(0, len(rear_df.index)-1, 300)
strindex_df = rear_df.index.strftime("%d/%m/%Y")
xticklabel=[]
for i in range(0,len(rear_df.index),10):
xticklabel.append(strindex_df[i])
for j in range(2):
for i in range(5):
if(j==0):
inter.append(make_interp_spline(list(range(len(rear_df.index))), rear_df.iloc[:,i]))
y_new.append(inter[i](x_new))
axe[i][j].plot(x_new,y_new[i], color='gray',label='Daily Cases')
axe[i][j].fill_between(x_new, y1, y_new[i], where=y_new[i]>=y1, facecolor=colour[i], interpolate=True, alpha=0.2)
axe[i][j].set_ylabel('Infected Cases',fontsize=20)
axe[i][j].set_xticks(list(range(0,len(rear_df.index),10)))
axe[i][j].set_xticklabels(xticklabel, rotation=30)
axe[i][j].set_title(cou[i], fontsize=20)
axe[i][j].legend(loc='best', bbox_to_anchor=(-0.2, 0.4, 0.5, 0.5),fontsize=15)
inter_cum.append(make_interp_spline(list(range(len(rear_df.index))), rear_df_cum.iloc[:,i]))
y_new_cum.append(inter_cum[i](x_new))
axe2 = axe[i][j].twinx()
axe2.plot(x_new,y_new_cum[i],'k--',linewidth=2,label='Total Cases')
axe2.legend(loc='best', bbox_to_anchor=(-0.2, 0.2, 0.5, 0.5),fontsize=15)
else:
inter.append(make_interp_spline(list(range(len(rear_df.index))), rear_df.iloc[:,i+5]))
y_new.append(inter[i+5](x_new))
axe[i][j].plot(x_new,y_new[i+5], color='gray', label= 'Daily Cases')
axe[i][j].fill_between(x_new, y1, y_new[i+5], where=y_new[i+5]>=y1, facecolor=colour[i+5], interpolate=True, alpha=0.2)
axe[i][j].set_ylabel('Infected Cases',fontsize=20)
axe[i][j].set_xticks(list(range(0,102,10)))
axe[i][j].set_xticklabels(xticklabel, rotation=30)
axe[i][j].set_title(cou[i+5], fontsize=20)
axe[i][j].legend(loc='best', bbox_to_anchor=(-0.2, 0.4, 0.5, 0.5),fontsize=15)
inter_cum.append(make_interp_spline(list(range(len(rear_df.index))), rear_df_cum.iloc[:,i+5]))
y_new_cum.append(inter_cum[i+5](x_new))
axe2 = axe[i][j].twinx()
axe2.plot(x_new,y_new_cum[i+5],'k--',linewidth=2, label='Total Cases')
axe2.legend(loc='best', bbox_to_anchor=(-0.2, 0.2, 0.5, 0.5),fontsize=15)
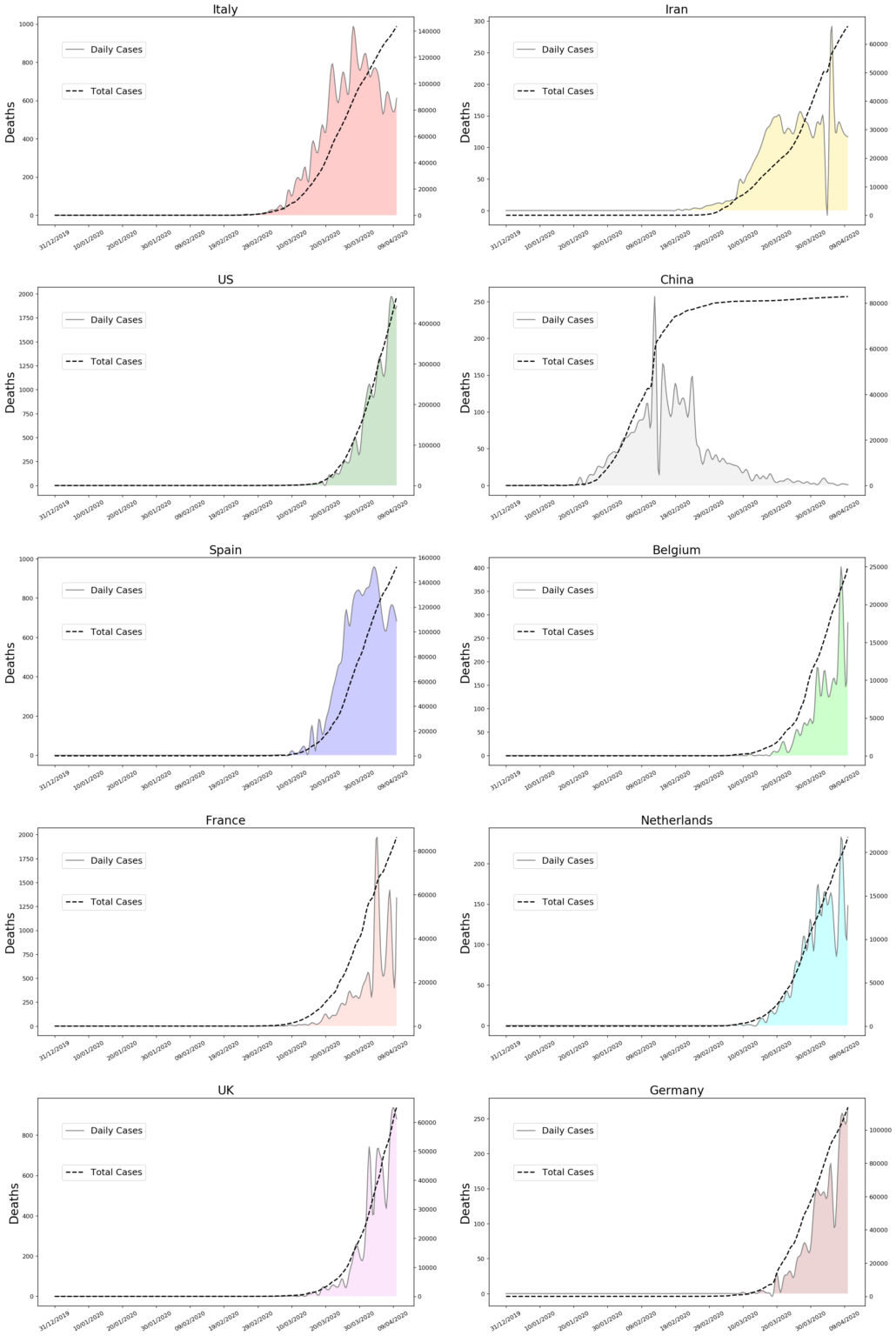
plt.savefig('infected_2.png',dpi=120,bbox_inches='tight')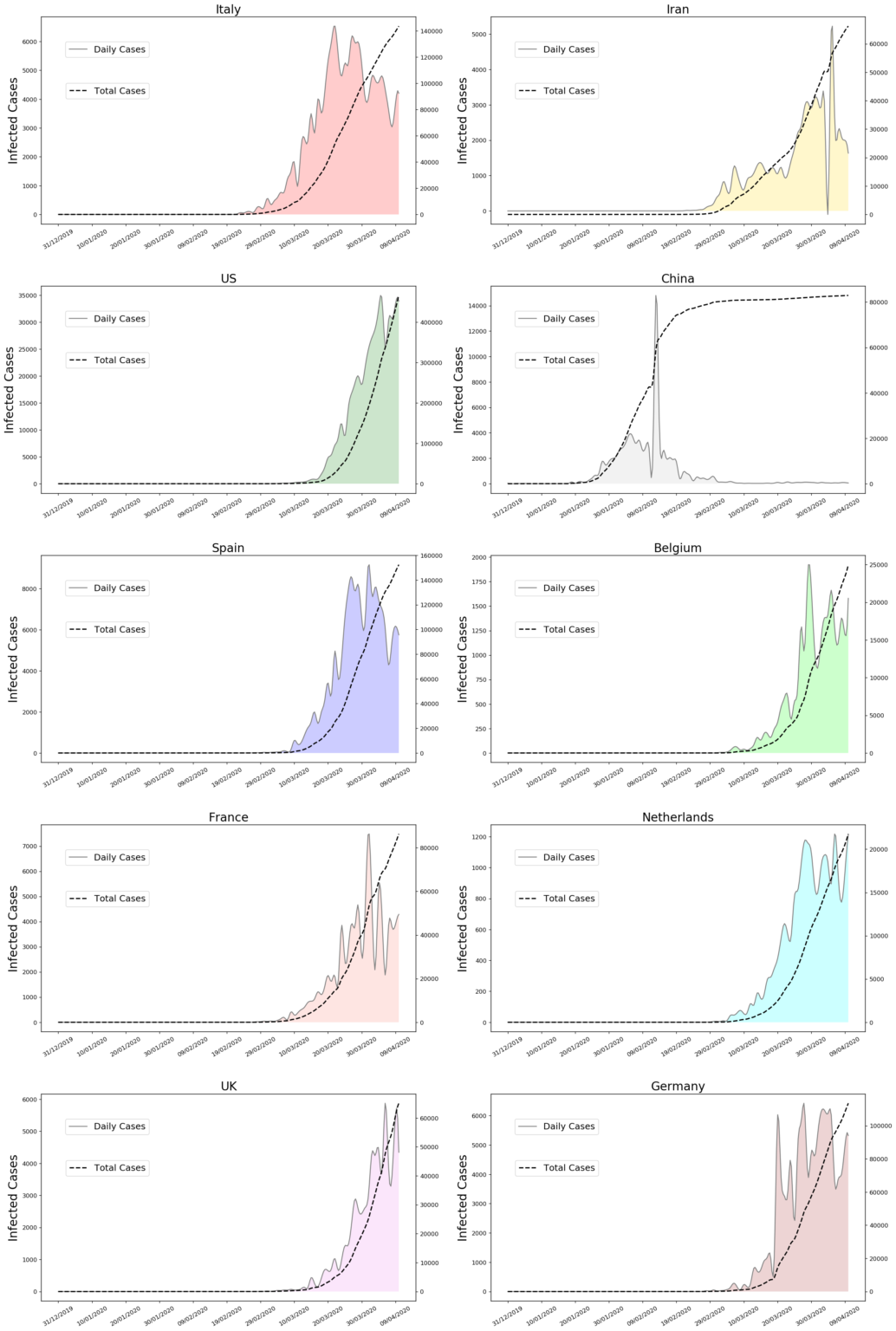
from matplotlib import pyplot as plt
import numpy as np
from scipy.interpolate import make_interp_spline
inter=[]
y_new=[]
inter_cum=[]
y_new_cum=[]
y1=0
colour = ['r','g','b','salmon','violet','gold','silver','lime','cyan','brown']
fig, axe = plt.subplots(5,2,figsize=(25,40))
plt.subplots_adjust(hspace=0.3)
x_new = np.linspace(0, len(rear_df.index)-1, 300)
strindex_df = rear_df.index.strftime("%d/%m/%Y")
xticklabel=[]
for i in range(0,len(rear_df.index),10):
xticklabel.append(strindex_df[i])
for j in range(2):
for i in range(5):
if(j==0):
inter.append(make_interp_spline(list(range(len(rear_df.index))), rear_df.iloc[:,i+10]))
y_new.append(inter[i](x_new))
axe[i][j].plot(x_new,y_new[i], color='gray',label='Daily Cases')
axe[i][j].fill_between(x_new, y1, y_new[i], where=y_new[i]>=y1, facecolor=colour[i], interpolate=True, alpha=0.2)
axe[i][j].set_ylabel('Deaths',fontsize=20)
axe[i][j].set_xticks(list(range(0,len(rear_df.index),10)))
axe[i][j].set_xticklabels(xticklabel, rotation=30)
axe[i][j].set_title(cou[i], fontsize=20)
axe[i][j].legend(loc='best', bbox_to_anchor=(-0.2, 0.4, 0.5, 0.5),fontsize=15)
inter_cum.append(make_interp_spline(list(range(len(rear_df.index))), rear_df_cum.iloc[:,i]))
y_new_cum.append(inter_cum[i](x_new))
axe2 = axe[i][j].twinx()
axe2.plot(x_new,y_new_cum[i],'k--',linewidth=2,label='Total Cases')
axe2.legend(loc='best', bbox_to_anchor=(-0.2, 0.2, 0.5, 0.5),fontsize=15)
else:
inter.append(make_interp_spline(list(range(len(rear_df.index))), rear_df.iloc[:,i+15]))
y_new.append(inter[i+5](x_new))
axe[i][j].plot(x_new,y_new[i+5], color='gray', label= 'Daily Cases')
axe[i][j].fill_between(x_new, y1, y_new[i+5], where=y_new[i+5]>=y1, facecolor=colour[i+5], interpolate=True, alpha=0.2)
axe[i][j].set_ylabel('Deaths',fontsize=20)
axe[i][j].set_xticks(list(range(0,len(rear_df.index),10)))
axe[i][j].set_xticklabels(xticklabel, rotation=30)
axe[i][j].set_title(cou[i+5], fontsize=20)
axe[i][j].legend(loc='best', bbox_to_anchor=(-0.2, 0.4, 0.5, 0.5),fontsize=15)
inter_cum.append(make_interp_spline(list(range(len(rear_df.index))), rear_df_cum.iloc[:,i+5]))
y_new_cum.append(inter_cum[i+5](x_new))
axe2 = axe[i][j].twinx()
axe2.plot(x_new,y_new_cum[i+5],'k--',linewidth=2, label='Total Cases')
axe2.legend(loc='best', bbox_to_anchor=(-0.2, 0.2, 0.5, 0.5),fontsize=15)
plt.savefig('deaths_2.png',dpi=120,bbox_inches='tight')